Onderzoeksmethoden 2/het werk/2012-13/Group 4
Inhoud
General information
Summary
For this assignment we are going to compare Information Science (IS) and Computing Science (CS) master's theses on for instance used research methods, use of language and length of sentences. We are doing this to answer the question: Is there a significant difference between IS and CS theses and is it possible to find classifiers to distinguish theses from these two related fields of study?
Problem statement
In Nijmegen only one Education Institute is responsible for the studies Computing Science and Information Science. Therefore there is a similarity between these two studies but both of them have their own specialization. Computing Science focusses on smart and secure systems, Information Science on architecture in the digital world.
One of the most important tasks of an education institute is testing the level of the students. The most important test of students in Nijmegen is the master thesis, which is written at the very end of their Master study.
The education institute, but also the The Accreditation Organisation of the Netherlands and Flanders (NVA), could ask whether there are significant differences between the two related studies. If there are differences, the next question will be what these differences are. Differences can be in meta data of the theses such as grade, writing time, supervisor or number of pages. But of course there will also be differences in the content of the theses. The meta data of the theses is most probably already gathered by the institute, but as far as we know there is no data about the content of the theses. So we will set up a research about the content of CS and IS master theses.
We would like to answer the question what the differences between CS and IS theses are. Therefore we are going to analyse the data with data mining techniques as described in the section data analysis, on the points described in the section research method.
This means we are going to try to answer this research question:
Is there a significant difference between IS and CS theses content?
As a subquestion we try to find a datamining classifier to classify theses on their content:
Is it possible to find classifiers to distinguish theses from these two related fields of study?
Research method
In order to compare theses we are going to look at the following parameters:
- Min, max and avg length of sentences.
- Size of vocabulary, minus commonly used words.
- Commonly used words which might indicate that the thesis comes from Information Science or Computer Science
The variables that we will check will be collected using a python script. The python script uses the NLTK library, which offers many functions for natural language processing.
We planned to look in to the different research methods that were used in the theses as well, but this turned out to be infeasible in the amount of time that we have for this course. This would have meant that we had to do a lot of manual checking in all theses or that we had to have a method to automatically classify the research methods. As this would take too much time and since we planned on doing a data analysis of all written words, we decided the used research method was not a part of this. We decided that this would be a good start of the chapter 'Future research' to see whether there is a significant difference between methods used by IS or CS students.
Conceptual model
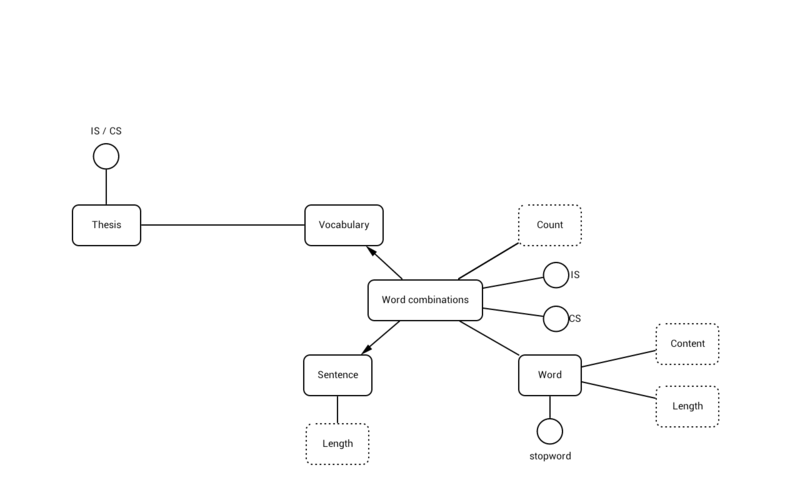
In the following model we see the values that are addressed in our Data Analysis:
On the left hand side we see the Thesis, which can either be an IS or CS thesis. This judgement is based on the vocabulary in the thesis. The vocabulary consists of word combinations. The word combinations is an abstract term for all words, either in a relation or separate. The actual word combinations in the literal sense are not processed in this research, but offer a good entry in the 'Future research', it will be interesting to see whether certain combinations are indications of a thesis being IS or CS.
The word combination 'sentence' is checked, and all sentence lengths in the thesis are stored. Words are then processed, and are checked for length and content. Length is the amount of letter is a word, content is the word itself. This is stored per thesis, and the frequency of the word occurring is stored.
Finally, stopwords like 'and', 'or' and other defined stopwords are not included in the analysis.
Data collection
The resource of theses will be the list of theses in the Master Thesis Lab of the Radboud institute for computer and information science[1]
A bundle of these files can be found at: http://schileffski.nl/uni/Masterscripties.zip
We have chosen to use an inductive analysis. We have gathered all master theses and worked from the bottom up. This allowed us to discover new concepts and theory about these concepts. Discovery of relationships between the concepts was our main goal for this research.
Beside this grounded theory, we also used some quantitative analysis. When counting all words, phrases and paragraphs, we abstracted from the meaning of all words. The richness of the language was not an important part in this analysis. Our choice was based on the fact that with qualitative data analysis, all data is hard to analyse in a systematic and objective way.
Data clean up
To make sure all data we wanted to analyse was useful, we started out with processing all Theses in PDF format, cleaning up the raw data to derive just the important information. We wanted to exclude all appendices, notes, codes, front pages and indexes. This would give us just the textual data. The first step in cleaning these PDF files was converting them to TXT files. TXT files are easier to edit and handle since the images and other PDF styles are not converted. After this conversion and clean up, the TXT files would be analyzed by the python script.
A problem with this clean up fase started with the conversion of the PDF files to TXT. When all 106 thesis were converted, it seemed all 'fi' letter combinations were converted to a period. This resulted in useless TXT files. The conversion had to be done again. After using a new program[2] for the conversion, first we made a check on one thesis to see if the conversion was correct. Since this was the case, we decided to make a distinction between the Information Science and Computer Science theses. We would run all PDF files through the converter and then clean the TXT files up by hand; deleting all unnecessary items. After all conversions were made, we starded cleaning up a few theses. We managed to clean up one thesis in around three hours time. Since this was a very short thesis, we calculated that doing all theses would leave us with approximately 320 hours of cleaning up. We decided to make a selection of the theses; 12 IS and 12 CS theses. This would still give us a view on the vocabulary and word use.
After cleaning up four theses the timer was already at 15 hours of cleaning up. This would take too much time. A new approach was decided. Each thesis was passed through the pdftotext linux commandline utility. Although this was not foolproof, it did generate usable files. These were then cleaned up by the script such that tokenizing on sentences and individual words, only words consisting of characters within the [a-zA-Z] range were left.
Because we did not remove the bibliographies from the textfiles, there were still some Dutch words to be found. After data reduction however, these were alle elliminated. Data reduction was done by calculating the cumulative frequencies of each attribute. After several trial runs with the datamining software and having issues handling the large dataset (30529 attributes), we made the decision to reduce the dataset such that only cumulative frequencies of 250 or greater were included. 34 percent of the initial dataset had a frequency count of 1, and 76 percent had a count of 10 or less. With the reduction to a cumulative frequency of 250, 633 attributes were left. Enough to run proper datamining algorithms. As to why not 249 of 251 as cutoff, there were a few different files made with various cutoffs (50,100,150,200,250,300,500) and it turned out that the 250 cutoff file performed quite well in the datamining algorithms. With full automation it might be possible to try every cutoff and see what performs best, but given time constraints and practical considerations the cutoff was set at the value of 250.
Data definition
We used 75 of the 106 theses to determine whether there is a significant difference between CS and IS studies. We deleted all theses written in Dutch which left us with 75 English written theses. Per study the most recent theses were used. From all 75 theses, 38 were from Computer Science and 37 were from Information Science. We wanted to the whole theses with the exception of the following parts:
- Title page
- Table of contents
- Source code of programs
- Appendices
- Tables
- Headers
- Page numbers
- Footers
In short, we wanted to use the flat text in natural language contained within the theses.
This however, was our initial idea. It proved to be hard to remove these objects from the texts as already decribed in the data cleanup section. In the end, most of the elements above were removed using the hard filter on just characters.
Data structuring
Every bit of code and other documents used in creating the dataset can be found at http://binarylotus.net/temp/tokenizerScriptsRM2.tar.gz (the file could not be uploaded on the Wiki because of filetype constraints).
The 75 remaining theses were fed to a tokenizer script that used the natural language toolkit (NLTK), a python library that allows for easy tokenization of texts. First, the entire text was split into sentences and the length of each sentence was determined. Then each sentence was tokenized into individual words and these were filtered on the [a-zA-Z] characterset:
<source lang="py">filter(lambda word: re.match("^[a-zA-Z]+$", word),sent)</source>
Besides the filter, these functions were important in the tokenizer script:
<source lang="py">
- takes the list and creates a wordList, using the filter described above.
- passes the senteces to the wordBag, which adds this word to the wordbag if it is not in there yet
def makeWordList(sentList):
wordList = []
for sent in sentList:
wordList.append(cleanupSentence(word_tokenize(sent)))
lengths.append(len(wordList[-1]))
wordBag(wordList[-1])
return wordList
def wordBag(sentence):
for word in sentence:
theWord = word.lower() #makes the word all lowercase
addToWordDict(theWord)
if theWord not in bagOfWords:
bagOfWords.append(theWord) #the bag of words is a collection of all words, so we know the size of the vocabulary used
def addToWordDict(word):
global wordCount
wordCount = wordCount+1 #Counting the number of words in the thesis
if word not in stopwords: #We gave a stopwordlist, so only words not in this list were added to the dictionary to become attributes
if word in wordDict:
wordDict[word] = wordDict[word]+1
else:
wordDict[word] = 1
</source>
The output of the script also contained various metrics on the thesis such as average sentence length, median sentence length and so on. These were simply calculated using the numpy library. The output of this script was a single CSV line, which grew longer with each thesis. Because all the lines in the CSV should contain an equal amount of attributes, a number of zero's would have to be appended such that each line would be equal in length to the last line in the CSV. This was done with a second script. Which also was used to create an arff style header. First, each line in the CSV should be read, which gave some trouble as not every element was an integer.
<source lang="py"> def parseLine(line):
line = line.split(',')
for elt in line:
elt = elt.rstrip(',')
elt = elt.lstrip()
line[0] = int(line[0])
line[1] = int(line[1])
line[2] = float(line[2])
line[3] = float(line[3])
for i in range(4, len(line)):
line[i] = int(line[i])
return line
</source>
After getting everything back into memory, it was a matter of appending zeroes to the length and then printing each line in the new array according to the CSV format.
The last step in the process was reducing the data according to a cutoff point, which we eventually set at a cumulative frequency of 250. To calculate these, we again had to load the CSV into memory, ignore the first eight attributes (as there were non-frequency attributes and the class attribute), transpose the matrix, calculate the sum, remove each line that did not match the cutoff, transpose again and stick it to the back of the first eight attribute-columns. This all was actually done in just two lines of code after setting things up:
<source lang="py"> my_data =numpy.array([col for col in my_data if sum(col) > cutoff]).T result = numpy.hstack((head_col.T,my_data)) </source>
We kept track of which attributes eventually made if through this script so that it was known which words these corresponded to. With everything in memory, it was a matter of printing everything to screen according to the format of an arff file. The writing of these scripts could have been done in a matter of hours by someone fluent in Python. We had only rudimentary understanding of this language to begin with and decided this project would be a great oppertunity to learn more about Python. Overall, more hours were spent on this than there were hours allocated to this course, mainly because of small quirck with the language and a lot of gotcha's that resulted in abandoning certain pieces of code and starting over. In the end there were results both for this project, and for ourselves with a better understanding of this language which has already proven useful for other courses as well.
Data analysis
The data analysis will be based on the output of the data gathering methods we will be using (the python scripts described above with the flat TXT files as input). From this data we shall try to derive patterns by using different data mining methods. The script produces a matrix with a frequency count for each unique word in the joint vocabularies of all the theses. Also, average sentence length, max sentence length, median sentence length, total word count, and vocabulary size are produced. Of course there is also an attribute that describes what sort of thesis it is: CS or IS. The result of the script (and the input for Rapid Miner) is a CSV-style file, cointaining a header that describes each attribute (being it a specific word, the average sentence length, the thesis sort etc.) and one line with all values for these attributes for each thesis. Most values will be "0" which means we are talking about a sparce matrix.
Choice of best classification algorithm
We compared the algorithms Bayes, k-NN and Rule Induction because they fit our dataset very well, as described in the algorithm descriptions below.
Description of algorithms
All descriptions of the algorithms are based on [3] and [4].
Naive Bayes
Naive Bayes (will be referenced as Bayes from now on) also works up to its name. This classification function works naive in such a sense, that it looks at all attributes of an object separately: it does not take dependencies between attributes in consideration. This means that for all attributes the function calculates the probability that this attribute belongs to a certain classification. After this has been done for all attributes, the total chance for the different classifications are calculated, and the classification with the highest chance is assigned.
For our project this suits very well, because all attributes among each other are independent. All attributes may contribute to the classification as much as the other attributes.
k-Nearest Neighbor
k-Nearest Neighbor (k-NN) has a name that pretty much basically describes what this classification function does. The function maps all objects to a multidimensional space (as much dimensions as there are properties), and simply assigns the classification that the majority of neighbors on all dimensions have. So the function, for each dimension, checks what classification the neighbor (so other objects with known classifications) objects have, calculates the classification that is most given to all these neighbors, and assigns this value to the object that is being classified.
The k in this function is the amount of nearest neighbors that are checked. If this value is for instance 3, then 3 neighbors will be checked on their classification.
This function is particularly useful in our research as all theses have a known classification. This means that all objects in the function can be mapped to all other objects, as the neighbor objects always have a classification available.
Rule Induction
The algorithm for Rule Induction that we used is a propositional rule learner named RIPPER. This algoritm starts with less prevalent classes and iteratively grows and prunes rules until there are no positive examples left or the error rate is greater than 50%. When growing, rules are added until the rule is perfect. The procedure tries every possible value of each attribute and selects the condition with the highest information gain (the amount that makes sure as much attributes as possible can be correctly assigned).
This works well with our dataset because it can work good on nominal values, and there is only a small amount of different classifications possible, which should keep the tree relatively small.
Description of evaluation
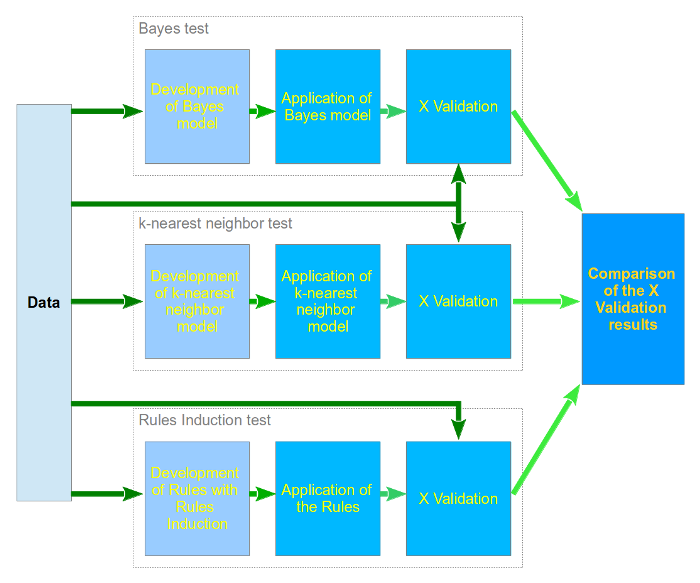
Our evaluation of the dataset is implemented as depicted in the model below:
As can be seen, the same data is used as input for all classification algorithms. The classification algorithms are run independently with all data in the dataset to create the first model. This model is then applied to the dataset, but in X fold validation.
X fold validation means that the dataset is chopped up in x same sized subsets, and per run one of the subsets is taken out as validation data for the model. The other X-1 subsets can then be used as training data on the model. The first set is then run through the adjusted model, and the score is saved. This is repeated X times, so that all subsets have been both training set and model set. When X runs have been done, the scores are averaged to create the final score of the model.
In this way the model is thoroughly tested and all objects have been training as well as validation data.
When all classification algorithms have been validated using the X fold validation, the end results will be compared to see which algorithm gives the best results on our dataset.
Results
All the three algorithms needed at most several minutes to classify the 75 theses with 633 attributes. So we think the efficiency of the three algorithms is at least sufficient and we compare only the average accuracy of every validation step of the three algorithms. The average accuracy is calculated with the X fold validation whereby X is the number of theses. So we validated every algorithm 75 times.
We also present the confusion matrices per algorithm, which show the total number of correct and incorrect classifications of all 75 validations. With the Bayes test for example 2835 theses (the total number of theses classified by the X fold validation) are classified correctly (99.47%) but 15 theses of Computer Science are classified incorrectly (as Information Science thesis). On the other hand 2540 theses of Information Science are classified correctly but 235 theses are incorrectly classified.
The total accuracy is calculated by making a weighted average of the IS and CS theses classification accuracy (because there is a difference in the amount of IS and CS theses).
Naive Bayes test
The bayes test ran without any further settings. The results are below in the matrix.
| True | Computer Science | Information Science |
|---|---|---|
| Computer Science | 2835 | 235 |
| Information Science | 15 | 2540 |
| Percentage | 99.47 % | 91.53% |
- accuracy: 95.56% +/- 0.73%
As we see here, the accuracy of the classification model that was built is quite fine, it ended up at 95.56%, with the CS theses being classified a lot better than the IS theses (99.47% versus 91.53%). The +/- 0.73% is the standard deviation on the results of the X fold validation.
k-NN test
For the k-NN classifier we used a setting of k = 1, which means that we only took the nearest neighbor in consideration for classifying the objects. We have run the classifier with higher k settings, but this gave results that were less accurate. Therefore k = 1 was chosen.
| True | Computer Science | Information Science |
|---|---|---|
| Computer Science | 2838 | 22 |
| Information Science | 12 | 2753 |
| Percentage | 99.58 % | 99.21% |
- accuracy: 99.40% +/- 0.66%
As we see here, the accuracy on the CS theses is slightly higher than with Naive Bayes, but significantly higher with the IS theses. The standard deviation of the X fold validation on the accuracy is only 0.66%. The overall accuracy is a lot higher than with Naive Bayes, which mostly comes from the better classified IS theses.
Rule Induction test
For the Rule Induction algorithm we used a desired pureness and sample ratio of 99%, which means that the algorithm will use at least 99% of the samples and will not stop until at least 99% of the objects is correctly classified. The minimal prune benefit is set to 25%, which means that the minimal benefit on a branch must be 25% in order not to be pruned.
| True | Computer Science | Information Science |
|---|---|---|
| Computer Science | 2833 | 12 |
| Information Science | 17 | 2763 |
| Percentage | 99.40 % | 99.57% |
- accuracy: 99.48% +/- 0.65%
This classification gives the best results. The CS theses are actually classified worst of all algorithms, with 0.07% less than the Bayes algorithm and 0.12% lower than the k-NN algoritm. However, the IS theses are classified best of all algorithms. The classification on these theses is 8.04% better than Naive Bayes, and 0,36% better than the k-NN algorithm. The standard deviation on the X fold validation results is the lowest of all classifiers, which together with the highest overall accuracy makes this the best classifier.
Chosen algorithm
We have chosen Rules Induction for the to-use best algorithm because it has the best accuracy and the model is very easy to interpret.
The Rules Induction with our whole data set results in the following rule model:
- if "link" ≤ 6.5 and "free" > 1.5 and "conclusion" > 7.5 then Computer Science (21 / 0)
- if "true" > 13.5 and "browser" ≤ 3.5 then Information Science (0 / 26)
- if "average" > 3.5 then Computer Science (9 / 0)
- if "employees" ≤ 5.5 then Information Science (0 / 8)
- if "procedure" ≤ 4 then Computer Science (8 / 0)
- else Information Science (0 / 2)
- correct: 100% of the training examples.
This means that for all 'if' statements a word attribute is meant (the attribute label shows which word this is, ie. the first word is "link") and the value for this word is shown (so the word count for the word "link" will have to be smaller or equal to 6.500).
When using the model above, all of the theses available can be classified correctly, giving a 100% classification result.
Conclusion
With these results we can get to the conclusion of our research. In this conclusion we shall give the answers to the main research question and the subquestion we proposed.
Is there a significant difference between IS and CS theses content?
Yes there is. There were quite a lot of words that occured less or more in the CS theses than in the IS theses (i.e. link, free, conclusion, average), and the other way round (i.e. true, browser, employees). For as far as we went in to the 'content' of the theses we can therefore state that this is true. Of course there is always the option to get in to the content a lot further, but that was not possible in this amount of time.
As with the primary research question, the answer is yes. We have been able to set up three classification algorithms that give results on our dataset that are near the 100%. One of the classifiers even gives a total of 100% of all instances classified correctly. This is a great result, and we are very happy that we got this. But there is a downside: we do not know for sure what will happen if new theses are entered in to the dataset. When we look at the final algorithm we chose, that gave us a 100% score, we see that the words that make up the rules do not seem to be very specific words that can be used to determine whether or not a thesis is a CS or IS thesis. However, it does work for 75 theses of each sort and the accuracy tested by X vault validation is very high, so it must say something. This will be an interesting part in future research, where there is more time and resources to check whether this model holds for more theses, or that it is just set up to work on this (relatively) small set of theses.
Future research
As mentioned before, there is quite some future research that can be done in this field. The points that we see fit for future research projects are:
- Researching whether the used research methods in the theses' research have any influence on the classification of the theses. This seems like a logical step, as there might be quite some difference in research methods used by IS and CS students.
- Researching whether combinations of specific words have influence on the classification of the theses. Looking in to the combinations of words that occur in the theses can be an adequate way to distinguish the two fields of study.
- Researching whether the created model is useful to classify new theses. This will ensure that the model is usable, and if it is not, it will show what needs to be changed in the model to make it more future proof.
- After a talk with Prof. Heskes, another idea was born. Not to look at the words in the thesis, but at so called dyads. Wherein each letter pair [i,i+1] is counted, resulting in a 26^2 matrix.
References
- ↑ Master Thesis Lab. (2012). Archive of Master Thesis Reports. http://www.ru.nl/iii/@761002/pagina/ [Online, accessed 23-09-2012]
- ↑ A-PDF. (2012). Thank your for your downloading ... A-PDF Text Extractor. http://www.a-pdf.com/text/download.htm [Online, accessed 15-10-2012]
- ↑ Han, J. , Kamber, M. (2006). Data Mining: Concepts and Techniques, Second Edition. Morgan Kaufmann.
- ↑ Hand, D., Manilla, H., Smyth, P. (2001). Principles of Data Mining. A Bradford Book.